Note: If you're interested in using it, feel free to ⭐️ the repo so we know!
The dataset can be downloaded from the link
The dataset can be downloaded from the link
A dataset that contains manually annotated 24,319 nuclei with associated class labels. The dataset can be downloaded from the link
The dataset consist of 143 images ER+ BCa images scanned at 40x. Each image is 2,000 x 2,000. Across these images there are about 12,000 nuclei manually segmented. The dataset can be downloaded from the link
Patch Generation has been done in offline mode to reduce pipeline complexity. The proposed approach uses multiple dataset therefore the size of WSIs are non-standard. Where as the pathches generated from WSIs have dimensions of 256x256x3 having 75% overlap among them.
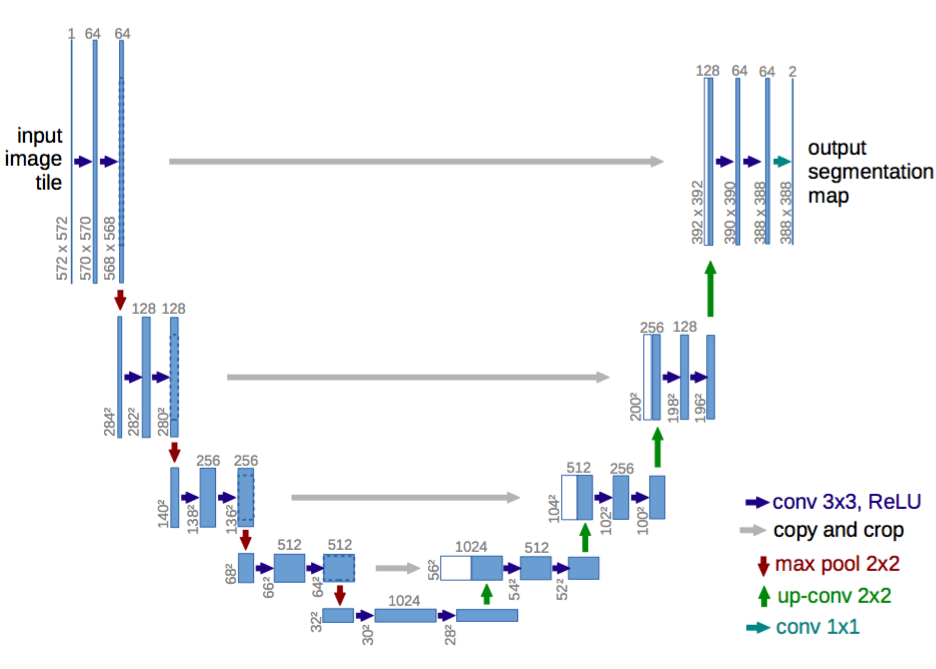
The purpose of this pipeline was to explore and train various nuclei segmentaion datasets therefore we used Modified U-Net for training.
The Pre-Trained models can be downloaded from google drive.
To get this repo work please install all the dependencies using the command below:
pip install -U segmentation-models
pip install -r requirments.txt
To start training run the Train.py script from the command below. For training configurations refer to the Training File file. You can update the file according to your training settings. Model avaible for training is U-NET.
python Train.py
To perfrom Inference on the trained models on Test Images you first have to download the weights and place them in the results folder. After downliading the weights you unzip them and then run the Inference by using the command below.
python Inference.py
Inferecne Results from CPM15 Dataset
| Tissue | Mask | Predicted Mask |
|---|---|---|
 |
 |
 |
Inferecne Results from CPM17 Dataset
| Tissue | Mask | Predicted Mask |
|---|---|---|
 |
 |
 |
Inferecne Results from Consep Dataset
| Tissue | Mask | Predicted Mask |
|---|---|---|
 |
 |
 |
Inferecne Results from Nuclei Segmentation Dataset
| Tissue | Mask | Predicted Mask |
|---|---|---|
 |
 |
 |
| Dataset | Loss | Accuracy | F1 Score | Dice Score |
|---|---|---|---|---|
| CPM 15 | 0.048 | 0.951 | 0.879 | 0.878 |
| CPM 17 | 0.054 | 0.945 | 0.866 | 0.865 |
| Consep | 0.304 | 0.692 | 0.586 | 0.586 |
| Nuclei Segmentation | 0.016 | 0.983 | N/A | N/A |
Model is evaluated on three metrics namely:
- Accuracy
- F1-Score
- Dice Score
Maintainer Syed Nauyan Rashid ([email protected])
Maintainer Asim Khan Niazi ([email protected])