2bit.el is a package for Emacs that provides a collection of functions,
macros and interactive commands that can be used to extract data from 2bit
format files. These are
files that can hold DNA sequences in a compressed format. You'll see samples
of this if you look at the human genome
data, for example.
The package is built such that it provides functions and macros for working with 2bit files, and then goes on to build interactive commands on top of this code. The elisp-level support code includes:
2bit-open should be called to "open" a 2bit file for further use:
(2bit-open FILE &optional MASKING)FILE is the path to the 2bit file you want to open. MASKING is an
optional parameter to say if mask blocks should be taken into account when
reading bases from the file. (it's worth noting that in my testing, mask
block handling tends to make the reading of data a lot slower)
2bit-sequence-count can be used to quickly get the count of how many
sequences are held in the 2bit file:
(2bit-sequence-count file)FILE can either be the path to a 2bit file, or a value returned from
2bit-open.
2bit-sequence-names can be used to quickly get a list of the names of all
the sequences held in a 2bit file:
(2bit-sequence-names file)FILE can either be the path to a 2bit file, or a value returned from
2bit-open.
2bit-sequence can be used to quickly get a named sequence from a 2bit
file:
(2bit-sequence file sequence)FILE can either be the path to a 2bit file, or a value returned from
2bit-open.
SEQUENCE is the name of the sequence to get.
2bit-sequence-dna-size returns the size of the DNA contained in the given
sequence.
(2bit-sequence-dna-size sequence)SEQUENCE must be a value returned from 2bit-sequence.
2bit-bases can be used to get a string of bases from a sequence in a 2bit
file:
(2bit-bases sequence start end)SEQUENCE is a value returned from a call to 2bit-sequence. START and
END describe the sub-sequence to grab. Note that the convention of
zero-based, inclusive of START and exclusive of END is used.
2bit-with-file is a simple macro provides as a convenience wrapper when
working with a 2bit file:
(2bit-with-file (handle file)
...body...)HANDLE is the name to give to the "handle" of the 2bit file, and FILE is
the file to open. For example:
;; Get a list of the sizes of each of the numbered chromosomes in the Human
;; Genome.
(2bit-with-file (hg "hg38.2bit")
(cl-loop for chr from 1 to 22
collect (2bit-sequence-dna-size (2bit-sequence hg (format "chr%d" chr)))))The following interactive commands are available:
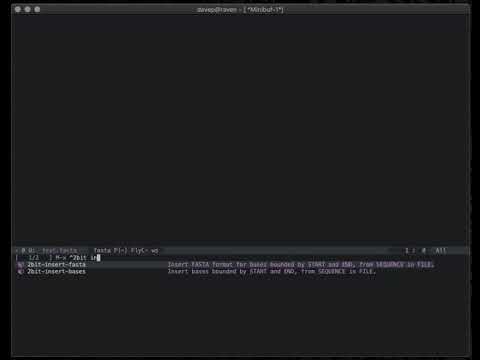
2bit-insert-bases simply inserts the requested sequence at the current
point in the current buffer.
2bit-insert-fasta simply inserts the requested sequence at the current
point in the current buffer, formatted in FASTA
format.
Here's a simple recording of a sample of using 2bit-insert-fasta to create
a FASTA file from a 2bit file: