I work for MeMed, a med-tech company developing a diagnostic tool based on host immune response.
We're using the host response to diagnose patients and reduce antibiotic overuse 🩺💊🦠 In my role as Scientific Affairs Lead I help establish international research collaborations, write papers about MeMed's latest clinical trial results and search for future funding opportunities
I did my PhD at Leibniz Institute of Photonic Technology, in Juergen Popp's group, working on research and development of innovative optical/photonic tools for point-of-care microbial diagnostics.
I used to teach at Friedrich Schiller Universität Jena - courses on Biophotonics and Raman spectroscopy
- The Pig That Wants to Be Eaten: 100 Experiments for the Armchair Philosopher by Julian Baggini
- Identity Crisis by Ben Elton
- I, Robot (Robot, #0.1) by Isaac Asimov
- The Godfather (The Godfather, #1) by Mario Puzo
Between 2019-2021, I worked on medical applications of Raman spectroscopy. The work was done with a wonderful team, under the supervision of Prof. Juergen Popp and Dr. Petra Roesch
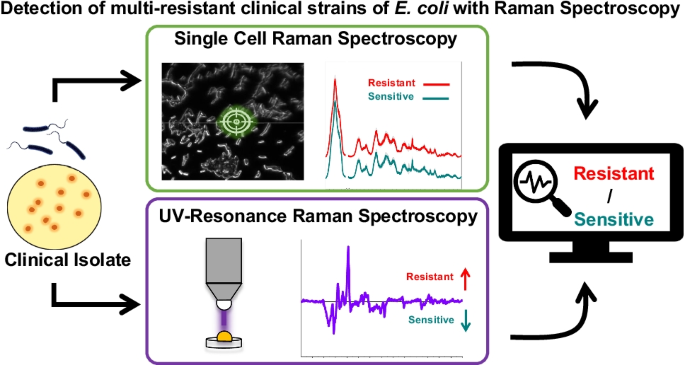
We were trying to use Raman spectroscopy for fast diagnostics of antibiotic resistant bacteria in hospitals.
In our work, we collected spectra directly from bacterial cells, and used machine learning algorithms (such as SVMs) to predict whether those bacteria are resistant to drugs. The idea --> to help doctors be able to prescribe the right drug 💊, to kill off the infection 🦠 - FAST!
- Nakar et al. 2022 - Detection of multi-resistant clinical strains of E. coli with Raman spectroscopy
- Nakar et al. 2022 - Label‐free Differentiation of clinical E. coli and Klebsiella isolates with Raman Spectroscopy
- Nakar et al. 2022 - Raman spectroscopy for the differentiation of Enterobacteriaceae: a comparison of two methods†
Enterobacteriacea | Klebsiella | E. coli Resistance | Sepsis | Raman Pipeline
- Raman Spectroscopic Method Diagnoses Infection at the Point of Care
- Invest in photonics now to fight infectious diseases and future pandemics
- Project Website
Between 2016-2018 I worked in Volcani Center. I was part of an interdisciplinary team which used fluorescence spectroscopy to detect water contaminations.
In that project, we collected water 💧 from different source wells and measured a huge spectrum 🌈 of auto-fluorescent signals. Using machine learning, we managed to identify the contaminations at X10 below the minimum required range, both in lab models and in our real collected water. That project was part of a collaboration with mekorot
Water |