First, let's download GraphBin-Tk. Make sure you have git installed.
git clone https://github.com/metagentools/gbintk.gitNow move in to the 'gbintk' directory.
cd gbintkLet's create an environment for gbintk using the provided environment.yml file. Make sure you have `conda`` installed.
conda env create -f environment.ymlActivate the environment.
conda activate gbintkTest your installation.
gbintk --helpNow we are all set for the demo.
Test data for the demo can be found in the tests/data/Sim-5G+metaSPAdes/ folder. Let's set the testing directory path to a variable as shown below so we don't have to type the path every time.
TESTDIR=tests/data/Sim-5G+metaSPAdes/Run the following command to bin the test dataset using MetaCoAG.
gbintk metacoag --assembler spades --graph $TESTDIR/assembly_graph_with_scaffolds.gfa --contigs $TESTDIR/contigs.fasta --paths $TESTDIR/contigs.paths --abundance $TESTDIR/coverm_mean_coverage.tsv --output $TESTDIRRun the following command to refine the binning results using GraphBin.
gbintk graphbin --assembler spades --graph $TESTDIR/assembly_graph_with_scaffolds.gfa --contigs $TESTDIR/contigs.fasta --paths $TESTDIR/contigs.paths --binned $TESTDIR/contig_to_bin.tsv --output $TESTDIRRun the following command to visualise the original binning result and the refined binning results on the assembly graph.
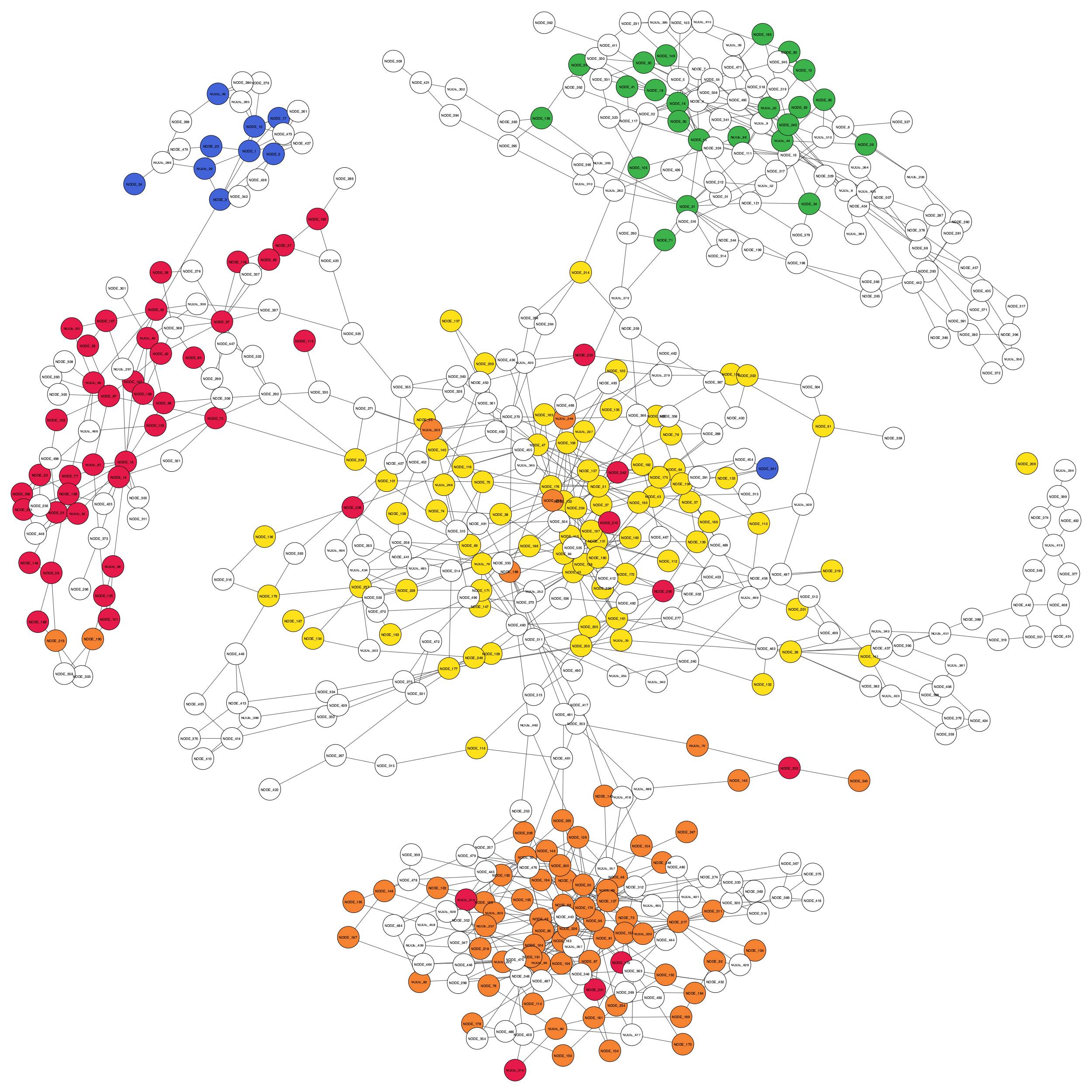
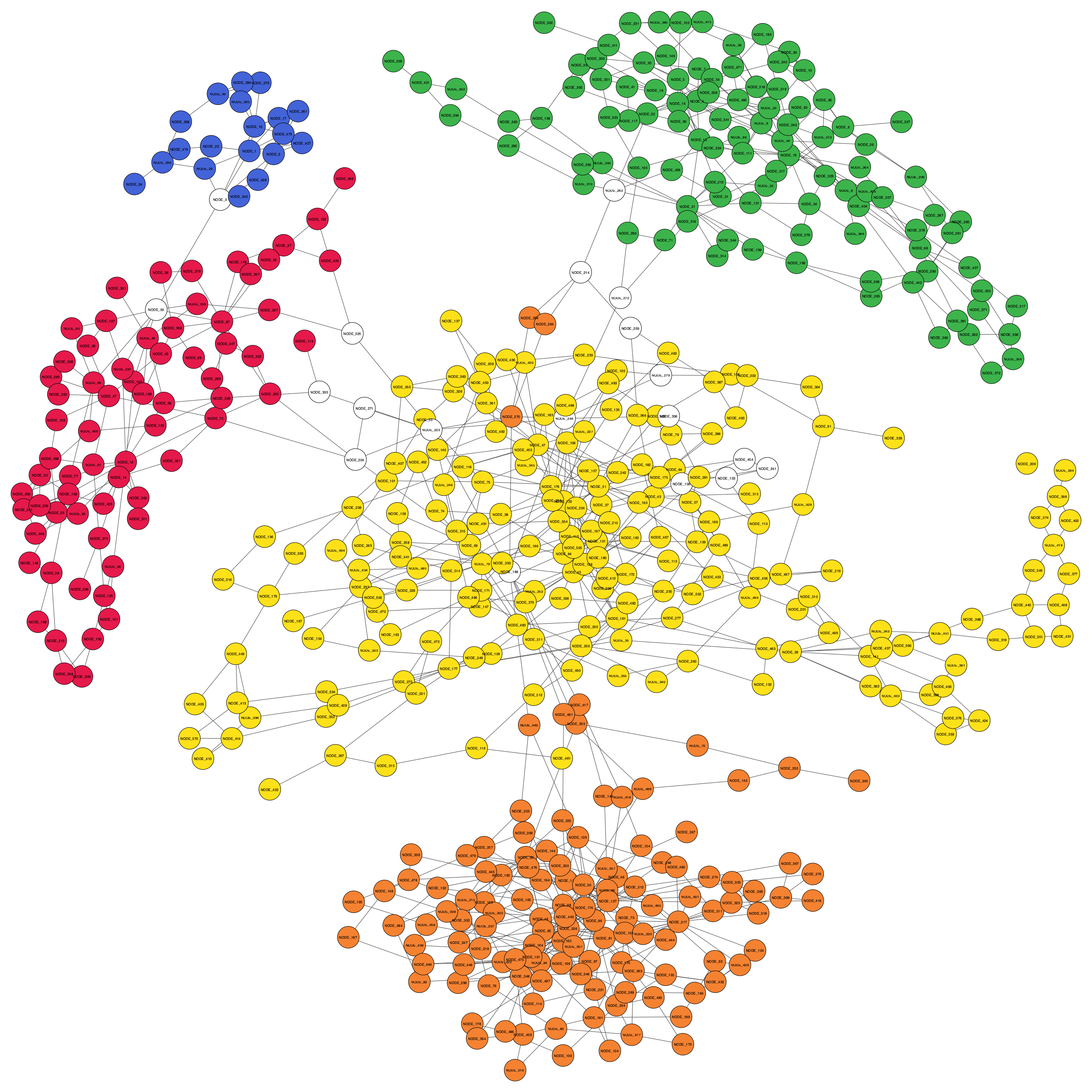
gbintk visualise --assembler spades --initial $TESTDIR/contig_to_bin.tsv --final $TESTDIR/graphbin_output.csv --graph $TESTDIR/assembly_graph_with_scaffolds.gfa --paths $TESTDIR/contigs.paths --output $TESTDIR --width 2500 --height 2500Below are some example visualisations generated.
Initial binning result
Refined binning result